Overall Gene Technology: Mind Map
Tip
Application based exam questions might ask you to apply a range of techniques, so it’s important you understand how they link together (e.g the sequence you could use them in, and the purpose of each one)
Spec Point
Spec Point
Recombinant DNA technology involves the transfer of fragments of DNA from one organism, or species, to another. Since the genetic code is universal, as are transcription and translation mechanisms, the transferred DNA can be translated within cells of the recipient (transgenic) organism.
As outlined in the spec point above, recombinant DNA technology involves the transfer of DNA fragments from one organism or species to another.
Recombinant DNA technology has many practical applications, such as: production of human insulin, growth hormone, pest-resistant GMO crops, or development of recombinant vaccine.
So how would bacteria be able to use human DNA to produce human proteins?
- The genetic code is universal
- This means that the same triplet codons code for the same amino acids in all species! So the codon AUG which codes for the amino acid Met, codes for this in bacteria and humans (all species)
- The mechanisms of transcription & translation are universal
- The overall mechanisms are the same across species, there are some differences in each organism’s regulation of the processes
Isolation of DNA fragments
Spec Point
Spec Point
Fragments of DNA can be produced by several methods, including: conversion of mRNA to complementary DNA (cDNA), using reverse transcriptase using restriction enzymes to cut a fragment containing the desired gene from DNA creating the gene in a ‘gene machine’.
Reverse Transcriptase
- An enzyme reverse transcriptase can make DNA from mRNA template - as the name suggests it’s the reverse of transcription (mRNA to DNA)
- We can use mRNA instead of DNA because mRNA has already been spliced and therefore does not contain introns (non coding DNA)
- Reverse transcriptase process
- Isolation of mRNA from desired gene of interest e.g Insulin from beta cells in pancreas
- Add DNA nucleotides and enzyme reverse transcriptase
- mRNA acts as template to form a single strand of cDNA (complementary DNA strand), using reverse transcriptase. This is due to complementary base pairing, see image below
- This single strand cDNA is then isolated (by removing the mRNA strand by hydrolysis or an enzyme)
- DNA polymerase is added to synthesise a double strand of DNA from cDNA
Restriction Endonucleases
Restriction endonucleases are enzymes found in bacteria. They recognise and cut DNA at specific DNA base sequences, known as recognition sites. Different restriction endonucleases can cut the DNA in different ways to:
-
Produce sticky ends* (This is the main one we focus on with in vivo cloning)
- This produces an uneven cut where 1 strand of DNA overhangs
- These overhangs can base pair with complementary DNA fragments
- These fragments can be joined by DNA ligase.
-
Produce blunt ends (Just to be aware that these enzymes can cut in different ways)
- These cut the DNA down the middle, leaving no DNA overhangs
What is a palindromic base sequence?
What is a palindromic base sequence?
A palindromic base sequence is a sequence of DNA that reads the same in opposite directions on the two complementary strands.
These sequences act as recognition sites for restriction endonucleases.
Example:
- 5’-GAATTC-3’
- 3’-CTTAAG-5’
Gene Machine
A gene machine is an automated device that synthesises DNA artificially, without needing a pre-existing DNA template.
The advantages of the gene machine:
- The DNA produced is intron-free, so it can be transcribed and translated by prokaryotic cells, which lack splicing machinery.
- Genes can be produced rapidly and accurately
- Gene machine process
- Identifies the amino acid sequence from the desired protein
- From this sequence the mRNA sequence and subsequent DNA sequence can be identified
- The gene machine synthesises short overlapping DNA fragments, called oligonucleotides.
- The oligonucleotides are joined together to form a longer DNA fragment, producing the complete gene
Amplification
Spec Point
Spec Point
Fragments of DNA can be amplified by in vitro and in vivo techniques.
Once the DNA fragment with the gene of interest has been isolated, we need to make a sufficient quantity of it. We can amplify the DNA in 2 ways:
- In Vivo - Transferring the fragments to host cell using a vector
- In Vitro- Polymerase Chain Reaction (PCR)
Note about the order in which to use different technologies
There may be instances where you want to amplify the DNA before isolating a fragment. For example, if there isn’t much of the target DNA you need to use, then you might do a PCR before using restriction endonucleases.
In Vitro PCR
Spec Point
Spec Point
The principles of the polymerase chain reaction (PCR) as an in vitro method to amplify DNA fragments.
Polymerase Chain Reaction (PCR), amplifies DNA, produces millions of copies of a specific fragment. It makes enough of the target DNA so it can be analyzed for genetic fingerprinting or mutations/diseases.
What's required for PCR?
What's required for PCR?
- Specific DNA fragment of interest
- Primers
- Short, specific base sequences of DNA
- Bind to complementary sequences on the DNA template
- Provide a starting point for DNA polymerase
- DNA polymerase (Taq polymerase)
- Derived from bacteria living in hot environments
- Is heat-stable and does not denature at high temperatures
- Nucleotides
- Used to build new DNA strands during replication
- PCR Steps
- Heat to 95°C to break hydrogen bonds between DNA template strand
- Reduce the temperature to 55°C to allow primers to bind to DNA
- Raise temperature to 72°C to allow DNA (taq) polymerase to join and add complementary nucleotides to DNA template. It begins at the primer until it reaches the end of the chain
Once this process is complete (and we have 2 strands of DNA), we then repeat the process with these 2 strands, resulting in 4 strands of DNA. As we keep repeating this process we can produce lots of new DNA strands very rapidly. The image below shows the exponential growth of the DNA strands (they double each round).
In Vivo Gene Cloning
Spec Point
Spec Point
The culture of transformed host cells as an in vivo method to amplify DNA fragments
- The addition of promoter and terminator regions to the fragments of DNA
- The use of restriction endonucleases and ligases to insert fragments of DNA into vectors. Transformation of host cells using these vectors
- The use of marker genes to detect genetically modified (GM) cells or organisms. (Students will not be required to recall specific marker genes in a written paper.)
In vivo gene cloning involves transferring the gene of interest into a host cell using a vector. This can produce the gene of interest in large quantities, e.g human insulin for production.
Isolation & preparation of DNA
To ensure the new host cells produce the proteins for the DNA fragment we are inserting, we need to add extra sequences of DNA. These include:
- Promoter - Binding site for RNA polymerase & transcription factors to allow initiation of transcription
- Terminator - Signal the end of transcription, providing a signal for RNA polymerase to detach from DNA template
Insertion into vector
The image below shows how we insert the DNA fragment into a vector. Vectors can then be used to transfer this DNA into host cells. There are different types of vectors, but in this example we are using plasmids found in bacteria.
- Insertion into vector steps
- Vector (plasmid) and DNA with gene of interest is cut at specific recognition sites using a restriction endonuclease enzyme
- Use the SAME restriction endonuclease to cut the DNA fragment (with gene of interest) and the plasmid. This ensures that the sticky ends of the opened plasmid are complementary to the sticky ends of the DNA (see image, step 2)
- The sticky ends attach to each other (via hydrogen bonding) and DNA ligase joins these fragments together, forming phosphodiester bonds in the sugar phosphate backbone.
- The plasmid now contains the new gene and is referred to as ‘recombinant DNA’
Transformation in host cell
- This recombinant plasmid now is reintroduced back into the bacterial cell
- The plasmids and bacterial cells are mixed together with calcium and heat. This makes it easier for the plasmids to pass through the cell membrane into cytoplasm of the bacterial cell
Marker Genes
Not all bacterial cells will take up the recombinant plasmid - it can be as few as 1%! Marker genes are used to identify the bacteria that have successfully taken up the gene. As the name suggests marker genes involve adding a separate gene on the plasmid - which makes the plasmid with the gene in identifiable. 3 examples below:
- Antibiotic resistance marker genes
- Fluorescence markers
- GFP (green fluorescent protein) gene
- Any bacteria that has taken up the gene will produce GFP
- Enzyme markers
- Gene that produces lactase
- Any bacteria that has taken up the gene will produce lactase
Tip
You don’t need to recall specific marker genes, but you should be able to understand how they work. Use diagrams/drawings to help you think through what is happening.
I’m going to outline how these marker genes work using the example of antibiotic resistance one.
Antibiotic resistance genes
- We can use a method called replica plating to see which plasmids have taken up the gene
- The image here shows:
- Bacteria cells without plasmids
- Plasmids without the gene taken up
- Plasmids with the gene taken up (recombinant plasmids)
What does the image show?
What does the image show?
Grown on ampicillin medium
Both plasmids have the genes for ampicillin resistance – so they can both grow in a medium of ampicillin. Note: if the bacteria doesn’t have any plasmids, it would not survive on this medium as the plasmid has the gene for ampicillin resistance.
Grown on tetracycline medium
Only the plasmids without the gene can grow. This is because the recombinant plasmids have their tetracycline gene cut by the restriction endonucleases as shown in the diagram.
Growth & Cloning
The identified bacteria (the ones with the gene of interest) are then cultured and divide by binary fission, producing many identical copies of the gene.
In Vivo vs In Vitro Comparison
| Feature | In vivo cloning | In vitro cloning |
|---|---|---|
| Definition | Cloning that occurs inside a living organism (usually a host cell) | Cloning carried out outside a living organism in a laboratory environment |
| Main enzymes involved | Host cell enzymes (e.g. DNA polymerase, DNA ligase) | Laboratory enzymes (e.g. DNA polymerase, restriction enzymes, ligase) |
| Typical vector | Plasmids or viral vectors | Usually no vector required |
| Speed | Relatively slow (depends on cell growth) | Very fast (can amplify DNA in hours) |
| Amount of DNA produced | Moderate to large amounts over time | Very large amounts in a short time |
| Control over conditions | Less control (depends on the host’s cellular processes) | High control (temperature, cycles, reagents) |
| Common uses | Gene cloning, protein production | DNA amplification for analysis, diagnostics |
Exam Question Practice
Describe how restriction endonuclease and DNA ligase are used to insert a gene into a plasmid.
(2 marks)Answer
Mark Scheme
- A restriction endonuclease cuts the plasmid, producing sticky ends (1 mark)
- Ligase joins gene to the plasmid by joining the sticky ends/forming phosphodiester bonds (1 mark)
Explain the shape of the curve in Figure 9.
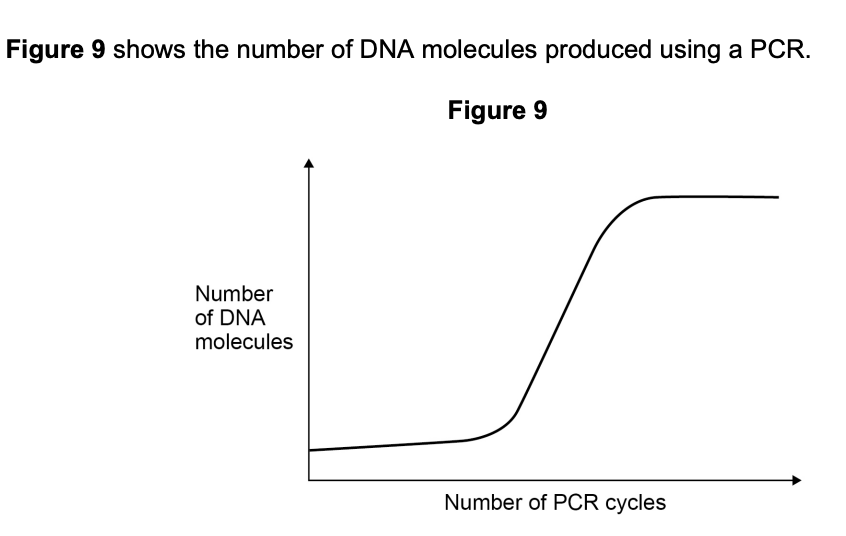 (2 marks)
(2 marks)
Answer
Mark Scheme
- Initially, the number of DNA molecules doubles each cycle, producing an exponential increase (1 mark)
- The curve plateaus when reagents such as nucleotides or primers become limiting (1 mark)
Describe and explain how the polymerase chain reaction (PCR) is used to amplify a DNA fragment.
(4 marks)Answer
Mark Scheme
- (Requires DNA fragment) DNA polymerase, (DNA) nucleotides and primers (1 mark)
- Heat to 95°C to break hydrogen bonds (and separate strands) (1 mark)
- Reduce temperature so primers bind to DNA/strands (1 mark)
- Increase temperature, DNA polymerase joins nucleotides (and repeat method) (1 mark)
Comparison of mitochondrial genes indicated that the diurnal geckos formed a distinct genetic group. This comparison also confirmed that all the geckos in the habitat were of the same species.
Explain how comparison of mitochondrial genes could indicate that the nocturnal geckos formed a distinct genetic group. In your answer, explain how new techniques enable the comparison of genes to be completed rapidly.
(3 marks)Hint
Think about what makes a genetic group distinct (different alleles/sequences) and name a technique that speeds up DNA comparison.
Answer
Mark Scheme
- Compare DNA base/nucleotide sequences OR banding patterns/positions of DNA fragments (1 mark)
- A distinct group will have different alleles/DNA sequences/banding patterns compared with other groups (1 mark)
- Use of a rapid technique (e.g. automated sequencing, PCR, electrophoresis, DNA probes) (1 mark)
Comments from mark scheme
- Common mistakes:
- Saying the geckos had different genes or were different species.
- Referring to mRNA or amino acid sequences instead of DNA.
- Saying “compare sequences” without stating DNA.
- Naming a technique without explaining how it is rapid.
- Failing to link different alleles/DNA sequences to genetic distinctness.
Comments from mark scheme