VNTRs
Spec Point
Spec Point
An organism’s genome contains many variable number tandem repeats (VNTRs). The probability of two individuals having the same VNTRs is very low.
The genome of most eukaryotic organisms contain many repetitive non-coding sequences of DNA, these are called Variable Number Tandem Repeat (VNTRs).
Individuals have a unique VNTR sequence (except identical twins) and the probability of any two people having the same VNTRs is extremely low. This property makes VNTRs excellent for genetic fingerprinting, where we want to distinguish specific individuals.
For example, the repeated sequence ATTG, I may have 30 copies of this, but my friend might have 1000 copies of this, and the barista at the coffee shop (where I got my coffee while writing this) may have 200 copies of this.
(Note: This can be any repeated sequence, it isn’t just ATTG)
What's the difference between VNTRs and introns?
What's the difference between VNTRs and introns?
- Introns are also non-coding sections within a gene. They are transcribed into pre-mRNA but removed during RNA splicing, so they are not translated into protein.
- VNTRs: Short, repetitive DNA sequences where the number of repeats varies between individuals, so they are useful for DNA profiling.
Note: VNTRs are often found in non-coding regions and can be located within introns or between genes (not always inside introns).
Gel Electrophoresis
Spec Point
Spec Point
The technique of genetic fingerprinting in analysing DNA fragments that have been cloned by PCR, and its use in determining genetic relationships and in determining the genetic variability within a population.
After amplifying the DNA through PCR, the results can then be visualised and analysed by using gel electrophoresis.
Gel electrophoresis
Is used to separate DNA fragments according to their size (number of nucleotides).
DNA fragments are pulled through a gel matrix (agarose) by an electric current. This separates DNA fragments according to the size (number of nucleotides) because:
- DNA fragments are negatively charged so move towards the positive electrode
- Smaller fragments move faster and travel further through the gel
We can compare size of the DNA fragments with a reference (DNA ladder) containing fragments of a known size.
Gel Electrophoresis Method
- Extract the DNA sample (e.g. from a hair strand)
- Increase the amount of DNA with PCR (This ensures there are enough VNTRs to analyse)
- Use restriction endonuclease to cut DNA into fragments, separating the VNTRs
- DNA Hybridisation. DNA probes are added to fragments. The probes have base sequences that are complementary to VNTRs so they can bind to target DNA sequence. Note: this is the same process as DNA probes
- Identification/development Use an X ray film to reveal the position of the DNA probes and the banding pattern
-Using Figure 1, which person has the longest DNA fragment?
-Using Figure 1, which person has the longest DNA fragment?
Person 1 (P1) as you can see the line is at 80bp (base pairs in size). This is the highest out of all three people
-Using Figure 2, which suspect is guilty (i.e their DNA is at the crime scene)?
-Using Figure 2, which suspect is guilty (i.e their DNA is at the crime scene)?
Suspect 2. The pattern of bands from the crime scene matches suspect 2
Genetic Fingerprinting Applications
Spec Point
Spec Point
The use of genetic fingerprinting in the fields of forensic science, medical diagnosis, animal and plant breeding.
Genetic fingerprinting can have many applications, including:
- Forensic science
- DNA (specifically VNTRs) left at a crime scene can be analysed and compared with the VNTR profiles of suspects.
- A match indicates the DNA likely came from the same individual.
- Medical diagnosis
- Genetic fingerprinting can be used to identify inherited genetic disorders.
- For example, Huntington’s disease, which involves a repeated nucleotide sequence (CAG) on chromosome 4.
- It can also be used to identify carriers of genetic conditions.
- Animal & plant breeding
- Genetic fingerprinting can be used to identify individuals with desirable alleles, such as:
- higher crop yield
- resistance to disease or environmental stress
- These animals or plants can then be selected for breeding to pass on the desirable traits
- Genetic fingerprinting can be used to identify individuals with desirable alleles, such as:
Exam Question Practice
BamHI and HindIII are both restriction endonucleases.
Figure 4 shows the positions where these enzymes cut a linear molecule of DNA.
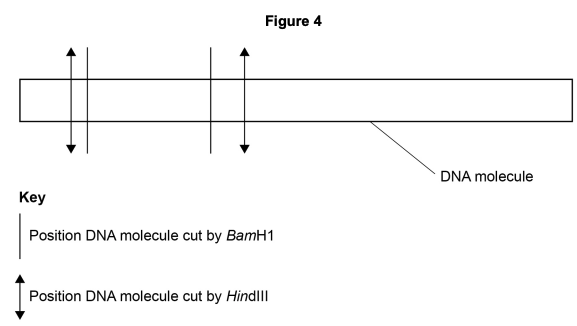
In two experiments, multiple copies of the DNA molecule shown in Figure 4 were all completely cut into fragments using these restriction enzymes. The DNA fragments produced were then separated by electrophoresis.
- Experiment 1 – DNA cut using BamHI
- Experiment 2 – DNA cut using BamHI and HindIII
Complete Figure 5 to show the relative positions of the bands following gel electrophoresis in experiments 1 and 2.
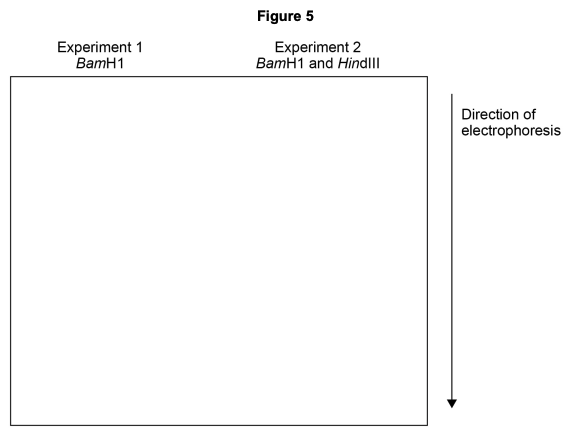 (2 marks)
(2 marks)
Hint
Work out how many fragments each experiment produces, then consider which fragments are longer (travel less distance).
Answer
Mark Scheme
Step 1: Work through how many fragments each experiment would produce. It is highly recommended to draw these out (see example below).
Step 2: Add the relative positions of these fragments to the gel electrophoresis diagram. Remember that longer fragments remain higher on the gel because they migrate more slowly.
Tips from examiner reports
- This was a tricky question, with only 14% of students answering it correctly. (2024 P2)
Suggest how you could determine the size of the different DNA fragments produced in these experiments.
(2 marks)Answer
Mark Scheme
- Use DNA fragments/ladder of known sizes/length (1 mark)
- Compare position/distance/bands with unknown fragment (1 mark)
Tips from examiner reports
- You need to ensure you mention compare the DNA ladder/DNA fragments of known sizes with the unknown fragments.
How many DNA fragments would be produced in experiment 2 if the original DNA molecule was a plasmid?
(1 marks)Hint
A plasmid is circular DNA. How does the number of cuts relate to the number of fragments in circular vs linear DNA?
Answer
Mark Scheme
- Answer = 4 (1 mark)
A plasmid is circular DNA, therefore the number of cuts will equal the number of fragments. If you look at the image you can see there are 4 cuts and 4 fragments.
Short tandem repeats (STRs) are short sequences of DNA, usually 2 to 7 base pairs. STRs are repeated a number of times, one after another. For example, the STR D5S818 is made up of AGAT repeated 7 to 16 times. STRs are found throughout the whole genome. The repeated sequences in STRs are common to all humans. Due to variation in the number of repeats, STRs can be used in genetic fingerprinting.
Describe how STRs could be removed from a sample of DNA.
(2 marks)Answer
Mark Scheme
- Use restriction endonucleases/enzymes (1 mark)
- (Cut DNA) at specific base sequences/pairs OR (Cut DNA) at recognition/restriction sites (1 mark)
Tips from examiner reports
- For specific base sequences you could have said ‘palindromic sequences’.
Genetic fingerprinting using STRs requires amplification of the STRs using the polymerase chain reaction (PCR). The short base sequences either side of a specific STR are known.
Explain the importance of knowing these base sequences in PCR.
(2 marks)Hint
Think about what primers do in PCR and why their sequence matters.
Answer
Mark Scheme
- Knowing the base sequence allows scientists to make primers that are complementary to the DNA (1 mark)
- These primers provide a starting sequence for DNA/Taq polymerase (1 mark)
Tips from examiner reports
- This links in with the PCR topic.
Genetic testing, using DNA from saliva, can screen for all known harmful mutations in both genes. Describe how this DNA could be screened for all known harmful mutations in both genes.
(3 marks)Hint
Think about the steps: amplify → cut → separate → detect
- Use PCR to amplify the DNA so there is enough to analyse.
- Cut the DNA into fragments using restriction endonucleases.
- Separate DNA fragments using gel electrophoresis.
- Add labelled DNA probes that bind (hybridise) to complementary sequences associated with harmful mutations.
- Identify mutations using fluorescence or radioactivity, or by comparing band positions with known mutation patterns.
Answer
Mark Scheme
- Use PCR to amplify the DNA sample (1 mark)
- Cut DNA using restriction endonucleases (1 mark)
- Separate DNA fragments using gel electrophoresis (1 mark)
- Add labelled DNA probes which bind (hybridise) to complementary sequences (1 mark)
- Identify mutations by fluorescence/radioactivity OR by comparing band positions to known mutations (1 mark)
Comments from mark scheme